fastISM on DeepSEA Beluga¶
fastISM is a faster way to perform in-silico saturation mutagenesis. This tutorial uses the DeepSEA Beluga model (Zhou et al 2018), which predicts 2002 chromatin features for a 2000 bp input sequence. This tutorial covers the following:
- Installations and downloading required files for tutorial
- Benchmarking fastISM on the Beluga model against a standard ISM implementation
- Running fastISM on custom input sequences
- Visualizing fastISM output across all tasks (outputs)
- Selecting a task, visualizing the fastISM scores, and zooming in to visualize the underlying sequence features.
Installations and Data¶
We use pip to install fastISM and vizsequence (to visualize sequence importance scores). In addition we download a trained Beluga model and a tsv with all the model’s outputs.
[4]:
# install fastISM
!pip install fastism
Collecting fastism
Downloading https://files.pythonhosted.org/packages/4f/93/26f83f7197d92b0c502d7f7af32cbd5e0d0f0b52a4bfb51b29162e860fc8/fastism-0.4.0-py3-none-any.whl
Requirement already satisfied: tensorflow<3.0.0,>=2.3.0 in /usr/local/lib/python3.6/dist-packages (from fastism) (2.3.0)
Collecting pydot<2.0.0,>=1.4.1
Downloading https://files.pythonhosted.org/packages/33/d1/b1479a770f66d962f545c2101630ce1d5592d90cb4f083d38862e93d16d2/pydot-1.4.1-py2.py3-none-any.whl
Requirement already satisfied: numpy<1.19.0,>=1.16.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (1.18.5)
Requirement already satisfied: wrapt>=1.11.1 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (1.12.1)
Requirement already satisfied: scipy==1.4.1 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (1.4.1)
Requirement already satisfied: grpcio>=1.8.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (1.32.0)
Requirement already satisfied: six>=1.12.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (1.15.0)
Requirement already satisfied: keras-preprocessing<1.2,>=1.1.1 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (1.1.2)
Requirement already satisfied: astunparse==1.6.3 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (1.6.3)
Requirement already satisfied: absl-py>=0.7.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (0.10.0)
Requirement already satisfied: wheel>=0.26 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (0.35.1)
Requirement already satisfied: opt-einsum>=2.3.2 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (3.3.0)
Requirement already satisfied: google-pasta>=0.1.8 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (0.2.0)
Requirement already satisfied: gast==0.3.3 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (0.3.3)
Requirement already satisfied: protobuf>=3.9.2 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (3.12.4)
Requirement already satisfied: tensorboard<3,>=2.3.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (2.3.0)
Requirement already satisfied: tensorflow-estimator<2.4.0,>=2.3.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (2.3.0)
Requirement already satisfied: termcolor>=1.1.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (1.1.0)
Requirement already satisfied: h5py<2.11.0,>=2.10.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<3.0.0,>=2.3.0->fastism) (2.10.0)
Requirement already satisfied: pyparsing>=2.1.4 in /usr/local/lib/python3.6/dist-packages (from pydot<2.0.0,>=1.4.1->fastism) (2.4.7)
Requirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from protobuf>=3.9.2->tensorflow<3.0.0,>=2.3.0->fastism) (50.3.0)
Requirement already satisfied: tensorboard-plugin-wit>=1.6.0 in /usr/local/lib/python3.6/dist-packages (from tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (1.7.0)
Requirement already satisfied: google-auth-oauthlib<0.5,>=0.4.1 in /usr/local/lib/python3.6/dist-packages (from tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (0.4.1)
Requirement already satisfied: werkzeug>=0.11.15 in /usr/local/lib/python3.6/dist-packages (from tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (1.0.1)
Requirement already satisfied: markdown>=2.6.8 in /usr/local/lib/python3.6/dist-packages (from tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (3.2.2)
Requirement already satisfied: google-auth<2,>=1.6.3 in /usr/local/lib/python3.6/dist-packages (from tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (1.17.2)
Requirement already satisfied: requests<3,>=2.21.0 in /usr/local/lib/python3.6/dist-packages (from tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (2.23.0)
Requirement already satisfied: requests-oauthlib>=0.7.0 in /usr/local/lib/python3.6/dist-packages (from google-auth-oauthlib<0.5,>=0.4.1->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (1.3.0)
Requirement already satisfied: importlib-metadata; python_version < "3.8" in /usr/local/lib/python3.6/dist-packages (from markdown>=2.6.8->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (1.7.0)
Requirement already satisfied: cachetools<5.0,>=2.0.0 in /usr/local/lib/python3.6/dist-packages (from google-auth<2,>=1.6.3->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (4.1.1)
Requirement already satisfied: rsa<5,>=3.1.4; python_version >= "3" in /usr/local/lib/python3.6/dist-packages (from google-auth<2,>=1.6.3->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (4.6)
Requirement already satisfied: pyasn1-modules>=0.2.1 in /usr/local/lib/python3.6/dist-packages (from google-auth<2,>=1.6.3->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (0.2.8)
Requirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests<3,>=2.21.0->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (3.0.4)
Requirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests<3,>=2.21.0->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (2.10)
Requirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests<3,>=2.21.0->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (1.24.3)
Requirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests<3,>=2.21.0->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (2020.6.20)
Requirement already satisfied: oauthlib>=3.0.0 in /usr/local/lib/python3.6/dist-packages (from requests-oauthlib>=0.7.0->google-auth-oauthlib<0.5,>=0.4.1->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (3.1.0)
Requirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.6/dist-packages (from importlib-metadata; python_version < "3.8"->markdown>=2.6.8->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (3.1.0)
Requirement already satisfied: pyasn1>=0.1.3 in /usr/local/lib/python3.6/dist-packages (from rsa<5,>=3.1.4; python_version >= "3"->google-auth<2,>=1.6.3->tensorboard<3,>=2.3.0->tensorflow<3.0.0,>=2.3.0->fastism) (0.4.8)
Installing collected packages: pydot, fastism
Found existing installation: pydot 1.3.0
Uninstalling pydot-1.3.0:
Successfully uninstalled pydot-1.3.0
Successfully installed fastism-0.4.0 pydot-1.4.1
[ ]:
#for visualizing the per-position importance
!pip install vizsequence
Collecting vizsequence
Downloading https://files.pythonhosted.org/packages/a6/10/b3b210eba27de588fba3c261b80317413e18ac3e371df9578b3cdc61096c/vizsequence-0.1.1.0.tar.gz
Requirement already satisfied: numpy>=1.9 in /usr/local/lib/python3.6/dist-packages (from vizsequence) (1.18.5)
Requirement already satisfied: matplotlib>=2.2.2 in /usr/local/lib/python3.6/dist-packages (from vizsequence) (3.2.2)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=2.2.2->vizsequence) (2.4.7)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=2.2.2->vizsequence) (0.10.0)
Requirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=2.2.2->vizsequence) (2.8.1)
Requirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=2.2.2->vizsequence) (1.2.0)
Requirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from cycler>=0.10->matplotlib>=2.2.2->vizsequence) (1.15.0)
Building wheels for collected packages: vizsequence
Building wheel for vizsequence (setup.py) ... done
Created wheel for vizsequence: filename=vizsequence-0.1.1.0-cp36-none-any.whl size=3269 sha256=f5c7dd7708c4186bbdadb6e2e428969bec77fb97b43315a0eb60a4ddf1f6c62f
Stored in directory: /root/.cache/pip/wheels/08/eb/de/6b398b439ba39c278e5c341bdeed57d66280910e096496eaef
Successfully built vizsequence
Installing collected packages: vizsequence
Successfully installed vizsequence-0.1.1.0
[ ]:
# download trained model
! wget http://mitra.stanford.edu/kundaje/surag/fastISM/deepseabeluga_keras_nopermutelayer.h5 -O deepseabeluga.h5
--2020-09-20 06:25:21-- http://mitra.stanford.edu/kundaje/surag/fastISM/deepseabeluga_keras_nopermutelayer.h5
Resolving mitra.stanford.edu (mitra.stanford.edu)... 171.67.96.243
Connecting to mitra.stanford.edu (mitra.stanford.edu)|171.67.96.243|:80... connected.
HTTP request sent, awaiting response... 200 OK
Length: 598186116 (570M)
Saving to: ‘deepseabeluga.h5’
deepseabeluga.h5 100%[===================>] 570.47M 48.5MB/s in 12s
2020-09-20 06:25:33 (46.9 MB/s) - ‘deepseabeluga.h5’ saved [598186116/598186116]
[ ]:
# download output annotation
! wget https://raw.githubusercontent.com/FunctionLab/ExPecto/20b99d1278678/resources/deepsea_beluga_2002_features.tsv -O outputs.tsv
--2020-09-20 06:25:35-- https://raw.githubusercontent.com/FunctionLab/ExPecto/20b99d1278678/resources/deepsea_beluga_2002_features.tsv
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 151.101.0.133, 151.101.64.133, 151.101.128.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|151.101.0.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 203001 (198K) [text/plain]
Saving to: ‘outputs.tsv’
outputs.tsv 100%[===================>] 198.24K --.-KB/s in 0.04s
2020-09-20 06:25:36 (5.37 MB/s) - ‘outputs.tsv’ saved [203001/203001]
Init¶
[ ]:
import fastism
import tensorflow as tf
import numpy as np
from matplotlib import pyplot as plt
import pandas as pd
import seaborn as sns
import vizsequence
# for some seaborn warnings
import warnings; warnings.simplefilter('ignore')
import time
[ ]:
tf.__version__
'2.3.0'
[5]:
! pip freeze | grep fastism
fastism==0.4.0
[1]:
!nvidia-smi
Wed Sep 23 09:42:18 2020
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 450.66 Driver Version: 418.67 CUDA Version: 10.1 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 Tesla P100-PCIE... Off | 00000000:00:04.0 Off | 0 |
| N/A 34C P0 25W / 250W | 0MiB / 16280MiB | 0% Default |
| | | ERR! |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=============================================================================|
| No running processes found |
+-----------------------------------------------------------------------------+
[ ]:
print("Num GPUs Available: ", len(tf.config.experimental.list_physical_devices('GPU')))
Num GPUs Available: 1
[ ]:
device = 'GPU:0' if tf.config.experimental.list_physical_devices('GPU') else '/device:CPU:0'
device
'GPU:0'
Load Model¶
[ ]:
model = tf.keras.models.load_model("deepseabeluga.h5")
WARNING:tensorflow:No training configuration found in the save file, so the model was *not* compiled. Compile it manually.
[ ]:
model.summary()
Model: "sequential_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
conv1d_1 (Conv1D) (None, 1993, 320) 10560
_________________________________________________________________
activation_1 (Activation) (None, 1993, 320) 0
_________________________________________________________________
conv1d_2 (Conv1D) (None, 1986, 320) 819520
_________________________________________________________________
activation_2 (Activation) (None, 1986, 320) 0
_________________________________________________________________
dropout_1 (Dropout) (None, 1986, 320) 0
_________________________________________________________________
max_pooling1d_1 (MaxPooling1 (None, 496, 320) 0
_________________________________________________________________
conv1d_3 (Conv1D) (None, 489, 480) 1229280
_________________________________________________________________
activation_3 (Activation) (None, 489, 480) 0
_________________________________________________________________
conv1d_4 (Conv1D) (None, 482, 480) 1843680
_________________________________________________________________
activation_4 (Activation) (None, 482, 480) 0
_________________________________________________________________
dropout_2 (Dropout) (None, 482, 480) 0
_________________________________________________________________
max_pooling1d_2 (MaxPooling1 (None, 120, 480) 0
_________________________________________________________________
conv1d_5 (Conv1D) (None, 113, 640) 2458240
_________________________________________________________________
activation_5 (Activation) (None, 113, 640) 0
_________________________________________________________________
conv1d_6 (Conv1D) (None, 106, 640) 3277440
_________________________________________________________________
activation_6 (Activation) (None, 106, 640) 0
_________________________________________________________________
flatten_1 (Flatten) (None, 67840) 0
_________________________________________________________________
altereddense (Dense) (None, 2003) 135885523
_________________________________________________________________
activation_7 (Activation) (None, 2003) 0
_________________________________________________________________
dense_1 (Dense) (None, 2002) 4012008
_________________________________________________________________
activation_8 (Activation) (None, 2002) 0
=================================================================
Total params: 149,536,251
Trainable params: 149,536,251
Non-trainable params: 0
_________________________________________________________________
[ ]:
model.input.shape
TensorShape([None, 2000, 4])
[ ]:
# a look at the 2002 model outputs
outputs = pd.read_csv("./outputs.tsv", sep="\t")
outputs
| Unnamed: 0 | Cell type | Assay | Treatment | Assay type | Source | Unnamed: 6 | |
|---|---|---|---|---|---|---|---|
| 0 | 1 | 8988T | DNase | NaN | DNase | ENCODE | ./wgEncodeAwgDnaseUniform/wgEncodeAwgDnaseDuke... |
| 1 | 2 | AoSMC | DNase | NaN | DNase | ENCODE | ./wgEncodeAwgDnaseUniform/wgEncodeAwgDnaseDuke... |
| 2 | 3 | Chorion | DNase | NaN | DNase | ENCODE | ./wgEncodeAwgDnaseUniform/wgEncodeAwgDnaseDuke... |
| 3 | 4 | CLL | DNase | NaN | DNase | ENCODE | ./wgEncodeAwgDnaseUniform/wgEncodeAwgDnaseDuke... |
| 4 | 5 | Fibrobl | DNase | NaN | DNase | ENCODE | ./wgEncodeAwgDnaseUniform/wgEncodeAwgDnaseDuke... |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 1997 | 1998 | Osteoblasts | H3K4me2 | NaN | Histone | Roadmap Epigenomics | E129-H3K4me2.narrowPeak.gz |
| 1998 | 1999 | Osteoblasts | H3K4me3 | NaN | Histone | Roadmap Epigenomics | E129-H3K4me3.narrowPeak.gz |
| 1999 | 2000 | Osteoblasts | H3K79me2 | NaN | Histone | Roadmap Epigenomics | E129-H3K79me2.narrowPeak.gz |
| 2000 | 2001 | Osteoblasts | H3K9me3 | NaN | Histone | Roadmap Epigenomics | E129-H3K9me3.narrowPeak.gz |
| 2001 | 2002 | Osteoblasts | H4K20me1 | NaN | Histone | Roadmap Epigenomics | E129-H4K20me1.narrowPeak.gz |
2002 rows × 7 columns
Benchmark¶
Calculate approximate speedup of fastISM vs a naive implementation.
[ ]:
def time_ism(ism_model, batch_sizes, seqlen):
times = []
per_100 = []
for b in batch_sizes:
# using randomly initialized tensors for benchmarking
x = np.random.random((b,seqlen,4))
x = tf.constant(x, dtype=ism_model.model.input.dtype)
t = time.time()
# each position is substituted with 0s for benchmarking
ism_model(x)
times.append(time.time()-t)
per_100.append((times[-1]/b)*100)
print("BATCH: {}\tTIME: {:.2f}\tPER 100: {:.2f}".format(b, times[-1], (times[-1]/b)*100))
print("BEST PER 100: {:.2f}".format(min(per_100)))
[ ]:
fast_ism_model = fastism.FastISM(model, test_correctness=False)
[ ]:
# 512 works with 16Gb GPU memory
time_ism(fast_ism_model, [128, 512], 2000)
BATCH: 128 TIME: 42.58 PER 100: 33.26
BATCH: 512 TIME: 85.09 PER 100: 16.62
BEST PER 100: 16.62
[ ]:
naive_ism_model = fastism.NaiveISM(model)
[ ]:
time_ism(naive_ism_model, [128, 512], 2000)
BATCH: 128 TIME: 142.26 PER 100: 111.14
BATCH: 512 TIME: 513.30 PER 100: 100.25
BEST PER 100: 100.25
Naive ISM takes around 100 seconds per 100 sequence, while fastISM takes around 16.6 seconds. That’s a speedup of about 6x!
Run on Custom Sequences¶
We run fastISM on selected custom sequences, check correctness against a standard implementation and visualize the fastISM importance score matrix across the top 200 outputs.
[ ]:
# chr3:138862274-138864274 (hg38)
chr3_enhancer = "CCGGTGATTTTCTGGAGTCTATATCCTTCATCAGATTTTCCAAGGGGTGTCTGTCCCCTCAAAAGAATGATTGTCATTATTTGAAAGACTAGTTCCAGACAGATATTTTATACAAATTTTCCCAGCATTGACATCCCTGAACCAAACTGTTTTTCTTCCCAACATTACTGTTTTCTTCCTTTCTGTCGAGTTTGTTGTTTTGTAATATCAGAATCTCCAGCTCACCTGAGTAAATGGTAACAAGGTGCCACCACCTTTGAATTCTCCCAGAATCCACCCCACCCTCCGTCAGAGCCACTGCCAAGGCACTCTTACTGATTTCTCCCACACTGCTGGCTCATTGCAAGTGGGAAGACAGCATGTGGAGTGGGTGTGCGGCTTATTAAAGTGAGAACTCAGGGTCAGGGCAGAACCAGGAAGAGAGCAGTGAGATATCCTGCTACCTAATCCAATTCTCCTTTTTGTGCATTTAGCACCCTCCCCTCCGCCTGCATAACAATGGAAGGAAAGAGGAAGTGGGAAAAAAGAAAGTCATGTAATTGAGTTAGAAGAGGTAATGACCAAGACCCTGGAGCAGAGGGAAAGCGGGTTACAAAAGGTGGGTTAAAGAAATCACAAGAGTATGAAGAGCTGGGAAATTACTAACAAATATTTGCTTGTGTGGGAAAGCAAAAAAGTAAAAACTTCAGTGCTGAATTGGGGCGCTGAGCCACCAGGGAAATTTGAGATTGGCATCAAGGACCGTGTTGAAGCAGGGTGGGCGGAGAAGGAGGGAAAACTACCAGCCAGCTGAGATTTTGCAGCTAGGCTGTGGCCTGATACCGAGTATCGATGCCGCAAGGGAGGGATGAGTCAGTCCTAGCACGTCCAAGTTTAGAATAATAGACTGTTTGCCACTGGGAAGGCAAACACCTTTCCTGTGAGAGGGCTTGCTGACAGTTCCAATGTCCAAAGTCCAATGCCGACCCAGAAAACTGAGGAGGCCCTGGCCCCTGCAGGAAGGGCTCATTTACATGGAGACTGAGTAAAGTGCTGTCTTAAACCCTCCTTCCTTCCCCCACTGGGAGGTTTCAGCCAGATATGCCACCCTTTGTAGGATTTCATAGGGTTGTCTAAAGCCAGGGTTGGCACAGAGCAGAAGCCACAGGGCTAAGTACCAGATTATAATTGTCAATGTCACACCTTACTGCAGAAGCCAGGGAAGGGAGCTAGGAAACTGAAGAGCTTTCTTGGTTATGGGCGGGGCTGTAAATGCAGAGTGTGCCCTGGTGACTCATGGGAGACAGTGAGAAACACTGTGGGGATCTGGTCAACCGGGTACTGATTCCTTTGAGGAAGGTATACTCCACATGCCAACCTGATACTCATGGCTAGTGAAGAGATGGCAGGATTGGGTTGCATCAGCCAGCCTAACTCGACTTGGAAACACAGAAAATAACCCAGAGCAGGTCTCAAGCACTGTGTAACTTTATTAGTTCATAGTGGCTGAACAGCCATGTTTAGGGCCTCTCAGAAGAAAGAGTTTCATCTTTGGGAAGAAATTTGTGTTGGGTGATTTTGTTCATATAATTTTGTGTTTTTTGTTTTGTTTTGGTGTTTGAGACAGGGCCTCACTCTCTCACACAGGCTGGAGTGCAGTGGCACCATCTTAGCTCACTGCAACCTCTACCTTCCTGCCTCAAGCGATCCTCCTACTTCAGCCTCCTGCATAGCTGGGACTACAGGCACGTATCACTCAACCCAGCTAATTTTTTTTTTTTCGAGATGCAGTCTTGCTCTGTCACCCAGGCTGGAGAGCAATGGCACTATCTTGGCTCACTGTAACCCCCGCCTCCCAGTCTCTGCCTCCTGAGTAGCTGGGATTACAGGCTCCTGCCACCACCCCCGGCTCAGCTAATTATTTCTTTCTTTCTTTTTTCTGAGATGAAGTTTCACTCTTGTTGCCCAGGCTGGAGTGCAATGGCACGATCTCAGCTCACTGCAATGTCTGCTTCTGGGGT"
# reproduce to have a "batch" of sequences
# you can add more custom sequences to this list
sequences = [chr3_enhancer]*5
#We define a function to do the one-hot encoding
onehot_mapping = {
'A': [1,0,0,0],
'C': [0,1,0,0],
'G': [0,0,1,0],
'T': [0,0,0,1],
'N': [0,0,0,0],
'a': [1,0,0,0],
'c': [0,1,0,0],
'g': [0,0,1,0],
't': [0,0,0,1],
}
def one_hot_encode(sequence):
return np.array([onehot_mapping[x] for x in sequence])
onehot_sequences = np.array([one_hot_encode(x) for x in sequences])
onehot_sequences.shape
(5, 2000, 4)
[ ]:
x = tf.constant(onehot_sequences, dtype=model.input.dtype)
mutations = [[1,0,0,0],
[0,1,0,0],
[0,0,1,0],
[0,0,0,1]]
# get outputs with all mutations (A/C/G/T)
# in general, fastISM gives the most speedup for larger batch sizes
# here we use a small batch size for illustration
fast_ism_out = [fast_ism_model(x, replace_with=mut) for mut in mutations]
naive_ism_out = [naive_ism_model(x, replace_with=mut) for mut in mutations]
[ ]:
# make into a batch_size x seqlen x num_outputs x 4 (bases)
fast_ism_out = np.transpose(np.array(fast_ism_out), (1,2,3,0))
naive_ism_out = np.transpose(np.array(naive_ism_out), (1,2,3,0))
print(naive_ism_out.shape)
(5, 2000, 2002, 4)
[ ]:
# check correctness
print(np.allclose(fast_ism_out, naive_ism_out, atol=1e-6))
print(np.allclose(naive_ism_out, fast_ism_out, atol=1e-6))
True
True
[ ]:
# reference output (should return using ISM itself)
ref = model(x, training=False).numpy()
print(ref.shape)
# repeat to make shape compatible with ism_out (for broadcasting)
ref = np.expand_dims(np.repeat(np.expand_dims(ref, 1), 2000, 1), -1)
print(ref.shape)
(5, 2002)
(5, 2000, 2002, 1)
[ ]:
mut_minus_ref = fast_ism_out - ref
# euclidean distance from reference for each output at each position
mut_minus_ref_euclidean = np.sqrt(np.sum(np.square(mut_minus_ref), -1)/3)
print(mut_minus_ref_euclidean.shape)
(5, 2000, 2002)
[ ]:
# pick index of sequence to explore
SEQ_IDX = 0
[ ]:
# heatmap of importance scores [tasks (clustered) x position]
# for top 200 tasks by reference predictions
top_tasks = np.argsort(ref[SEQ_IDX][0][:,0])[::-1][:200]
to_plot = mut_minus_ref_euclidean[SEQ_IDX][:, top_tasks].T
cg = sns.clustermap(to_plot,
row_cluster=True, # cluster tasks
col_cluster=False, # not position
# standard_scale=0,
dendrogram_ratio=0.04,
figsize=(40,8),
xticklabels=100,
vmax = np.quantile(to_plot, 0.998), # set max limit
cbar_pos=(0, .3, .02, .4))
cg.ax_row_dendrogram.set_visible(False)
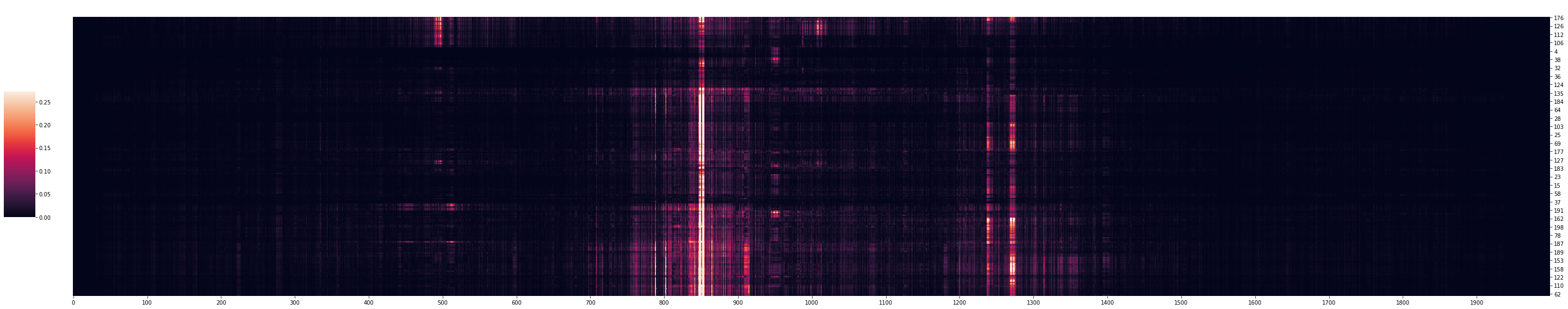
Positions in the input have different importance scores depending on the task. For example, regions around 500 bp have high importance scores for the top few tasks but not for the rest of the tasks.
Sequence Features for a Single Output¶
Let’s pick a task and zoom in to visualize the underlying sequence with high importance scores for that task.
[ ]:
# pick task with highest prediction
TASK_IDX = np.argsort(ref[SEQ_IDX][0][:,0])[::-1][0]
# or pick another task of your choice
# TASK_IDX = 1777
TASK_IDX
1777
[ ]:
# predicted value for that task
ref[SEQ_IDX, 0, TASK_IDX, 0]
0.98828876
[ ]:
# output (task) description
outputs.iloc[TASK_IDX]
Unnamed: 0 1778
Cell type Small_Intestine
Assay DNase.hot
Treatment NaN
Assay type DNase
Source Roadmap Epigenomics
Unnamed: 6 E109-DNase.hot.bed.gz
Name: 1777, dtype: object
The output is DNase-seq prediction in small intestine.
[ ]:
# heatmap of base x position difference from ref
plt.figure(figsize=(40,5))
to_plot = mut_minus_ref[SEQ_IDX, :, TASK_IDX].T
sns.heatmap(to_plot,
center=0,
vmin = np.quantile(to_plot, 0.0001),
vmax = np.quantile(to_plot, 0.9999),
xticklabels = 100,
yticklabels = ['A','C','G','T'],
cbar_kws = dict(use_gridspec=False,location="top"))
plt.show()
plt.figure(figsize=(40,2))
plt.plot(mut_minus_ref_euclidean[SEQ_IDX, :, TASK_IDX])
plt.margins(0, 0)
plt.show()
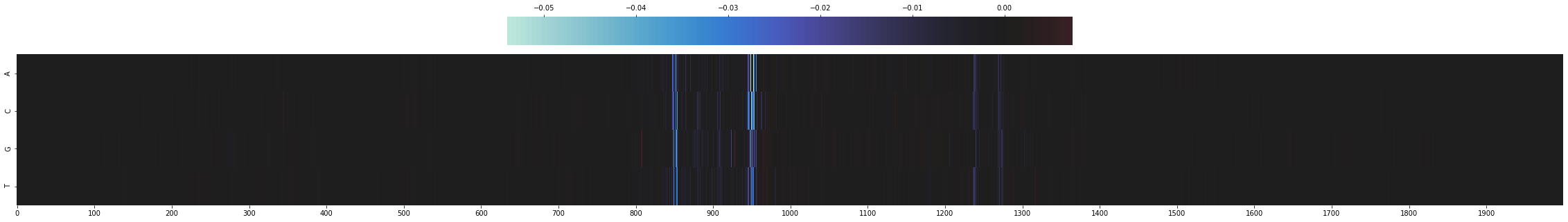

Let’s zoom into the central bases and visualize the sequence as well.
[ ]:
# Zoom in heatmap of base x position
ZOOM_IN = range(800, 1000)
plt.figure(figsize=(40,5))
to_plot = mut_minus_ref[SEQ_IDX, ZOOM_IN, TASK_IDX].T
sns.heatmap(to_plot,
center=0,
vmin = np.quantile(to_plot, 0.0001),
vmax = np.quantile(to_plot, 0.9999),
xticklabels = 10,
yticklabels = ['A','C','G','T'],
cbar_kws = dict(use_gridspec=False,location="top"))
plt.show()
plt.figure(figsize=(40,2))
plt.plot(mut_minus_ref_euclidean[SEQ_IDX, ZOOM_IN, TASK_IDX])
plt.margins(0.01, 0.01)
plt.show()
vizsequence.plot_weights(
np.expand_dims(mut_minus_ref_euclidean[SEQ_IDX, ZOOM_IN, TASK_IDX], -1)*onehot_sequences[SEQ_IDX, ZOOM_IN],
figsize=(40,2))
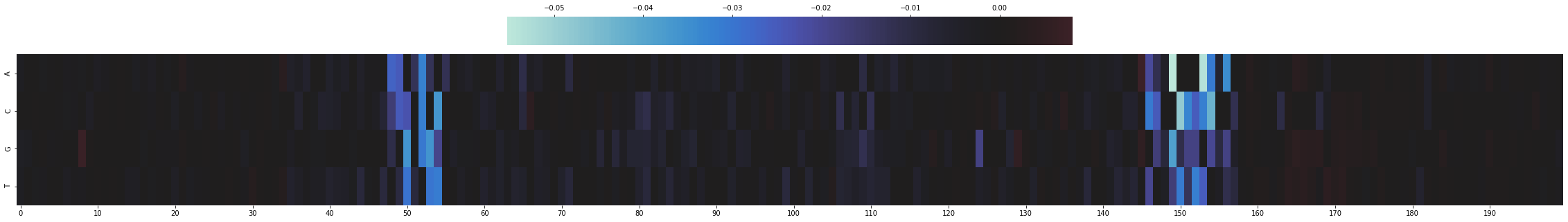


On the left is an AP1 motif and on the right is HNF4 motif. HNF4 has a known role in intestinal development.